Gini impurity & Entropy: Decision tree fundamentals
Is Gini impurity better that entropy
Quick recap: Entropy
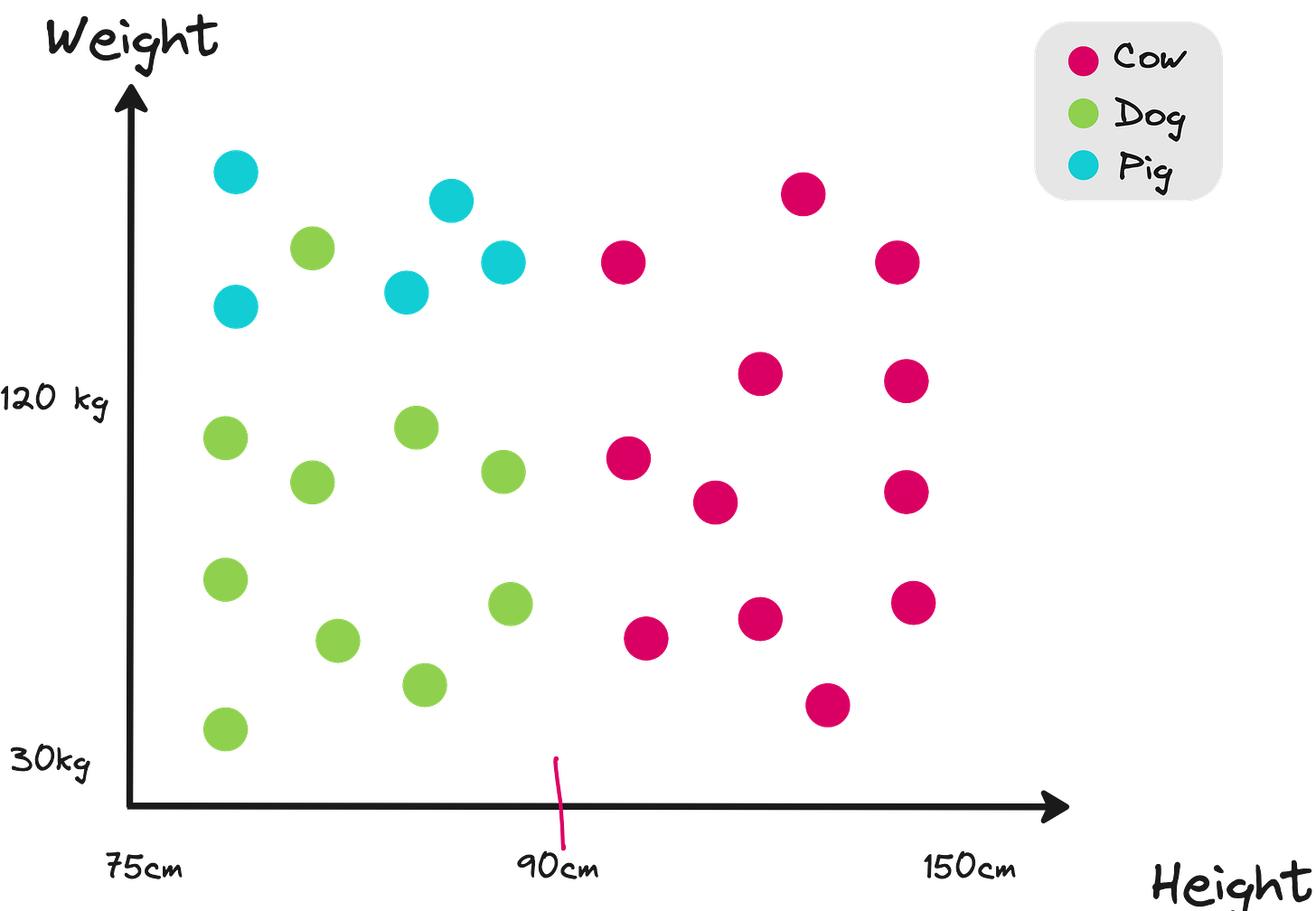
In this dataset, you want to classify a given animal to one of the 3 classes. Which feature would you use?
Height < 90cm or
Weight < 120kg?
You choose the feature that minimizes the weighted entropy of children nodes compared to the root node. So you go with the option in the left in the figure below.
Gini impurity introduction
Gini Impurity measures how often a randomly chosen element from a dataset would be incorrectly labeled if it was randomly labeled according to the distribution of labels.
If all data points belong to the same class in a node, the gini impurity = 0. These ndoes can be called as “pure leaves”.
Why is Gini impurity used?
It’s a measure of node impurity used in decision trees to determine node splits.
Lower Gini Impurity means better homogeneity (less impurity) within a node.
Comparison with other metrics:
Contrast Gini with Entropy (used in information gain).
Example: Gini is computationally simpler than Entropy.
Properties:
Perfectly pure split: Gini=0.
Completely random split: Gini>0.
Numerical example
Consider the below dataset with 1 feature with 2 values (red and blue) and 2 classes (yes or no). Calculate the gini impurity for parent node and the weighted impurity of children nodes.
Step 1: Initial node impurity
Step 2: Split the data
Step 3: Weighted Gini impurity
Step 4: Interpret results
After splitting, the weighted Gini impurity reduced from 0.5 to 0.444
Gini impurity v/s entropy
Gini impurity v/s entropy: When to use which?
Coding a node split and Gini impurity from scratch
Function for Gini Impurity
# Define a function to calculate Gini Impurity
function gini_impurity(data)
total = length(data)
if total == 0
return 0.0
end
# Count occurrences of each label
label_counts = Dict{String, Int}()
for row in data
label = row[:label]
label_counts[label] = get(label_counts, label, 0) + 1
end
# Calculate Gini Impurity
gini = 1.0
for count in values(label_counts)
prob = count / total
gini -= prob^2
end
return gini
endFunction for Splitting the Dataset
# Function to split data based on a feature value
function split_data(data, feature, value)
left = [row for row in data if row[feature] == value]
right = [row for row in data if row[feature] != value]
return left, right
endWeighted Gini Impurity Calculation
# Function to calculate weighted Gini Impurity for a split
function weighted_gini(data, splits)
total = length(data)
weighted_gini = 0.0
for split in splits
split_gini = gini_impurity(split)
weighted_gini += (length(split) / total) * split_gini
end
return weighted_gini
endExample Usage
# Sample dataset
data = [
Dict(:color => "Red", :label => "Yes"),
Dict(:color => "Red", :label => "Yes"),
Dict(:color => "Red", :label => "No"),
Dict(:color => "Blue", :label => "No"),
Dict(:color => "Blue", :label => "No"),
Dict(:color => "Blue", :label => "Yes")
]
# Calculate initial Gini Impurity
println("Initial Gini Impurity: ", gini_impurity(data))
# Split the data by the feature `color` with value `Red`
red_split, blue_split = split_data(data, :color, "Red")
println("Red Split: ", red_split)
println("Blue Split: ", blue_split)
# Calculate Gini Impurity after the split
split_gini = weighted_gini(data, [red_split, blue_split])
println("Gini Impurity after split: ", split_gini)